
Yong-Xin Liu, Professor at Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Executive Editor of iMeta. Research interesting are method and function in microbiome. He has published 80 papers in Science, Cell, Nature Biotechnology, Nature Microbiology, etc. These papers cited 29000+ times, and selected as the top 2% scientists and National Science Fund for Excellent Young Scholars. Founded a meta-genome WeChat account with 180,000+ followers, launched iMeta journal (IF 33.2), and ranked top 0.3% in global. Part-time reviewer for 100+ journals such as Cell Host & Microbes, Nature Communications, Nucleic Acids Research, and Microbiome > 360 times.
Research Experience
2022/10 - now Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences Professor / Principal Investigator
2021/1 - 2022/9 Institute of Genetics and Developmental Biology, Chinese Academy of Sciences Senior Engineer
2016/8 - 2020/12 Institute of Genetics and Developmental Biology, Chinese Academy of Sciences Engineer
2014/7 - 2016/7 Institute of Genetics and Developmental Biology, Chinese Academy of Sciences Postdoctoral Fellow
Education
2011/9-2014/6 University of Chinese Academy of Sciences Ph.D. of Bioinformatics, Supervised by Prof. Xiu-Jie Wang
2008/9-2011/6 Northeast Agricultural University Master of Crop Genetics and Breeding, Supervised by Prof. Wenbin Li
2004/9-2018/6 Northeast Agricultural University Bachelor of Biotechnology & Microbiology

Research Interest
Yong-Xin Liu's laboratory is dedicated to the methods development and functional mining of microbiome (Figure 1). The laboratory focuses on metagenomics/microbiome method development (Figure 2), with particular emphasis on new technologies such as third-generation and single-cell sequencing to elucidate the role of microbiota in agriculture, food, and human nutrition and health (Figure 3), and to build a science communication for the world (Figure 4).
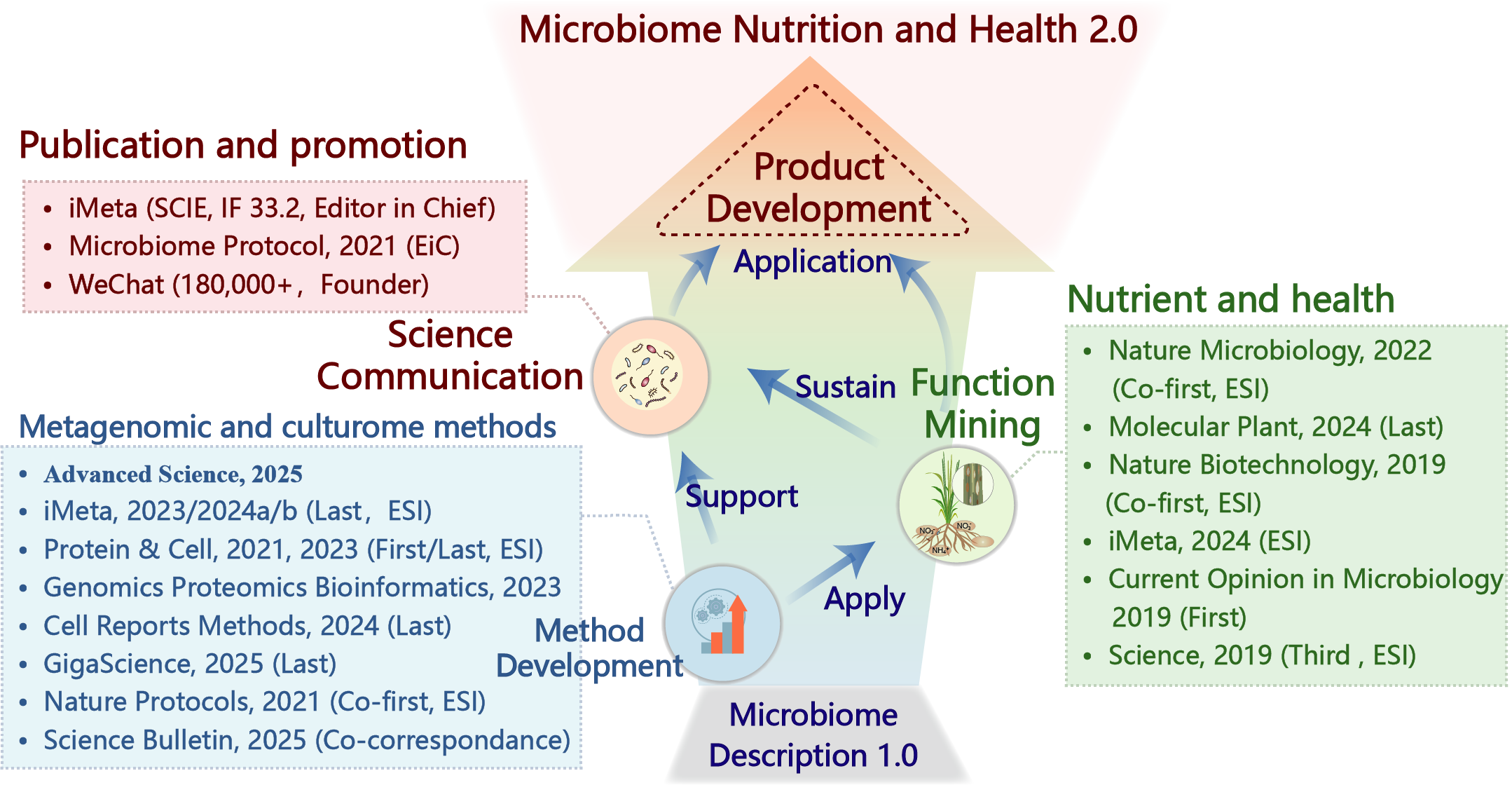
Figure 1. Method development and functional mining drive microbiome nutrition and health
1) Method Development: Efficient and user-friendly metagenomic data analysis and culturome for microbiomes were established. 2) Functional Mining: The above methods were applied to crop microbiome research, revealing new theories regarding the role of rice root microbiomes in nitrogen absorption, uncovering highly effective biocontrol bacteria against wheat scab and elucidating their mechanisms, elucidating a new mechanism by which polyphenols from traditional Chinese medicine alleviate enteritis through microbial metabolites, and discovering a new phenomenon linking vaginal microbiota with infertility, thus revealing the role of microbiomes in human and crop nutrition and health from multiple perspectives.
3) Publication and Promotion: A metagenomics WeChat account with over 180,000 followers, a microbiome protocol eBook was launched, and the world's top-ranked microbiology research journal, iMeta (IF 33.2), was founded, filling the gap in high-level publication and promotion in this field in Asia. Sub-journals such as iMetaOmics and iMetaMed have been established, with the goal of substitution for the Cell series. These achievements have accelerated the transformation of microbiome science from the species description 1.0 era to the nutrition and health 2.0 era.
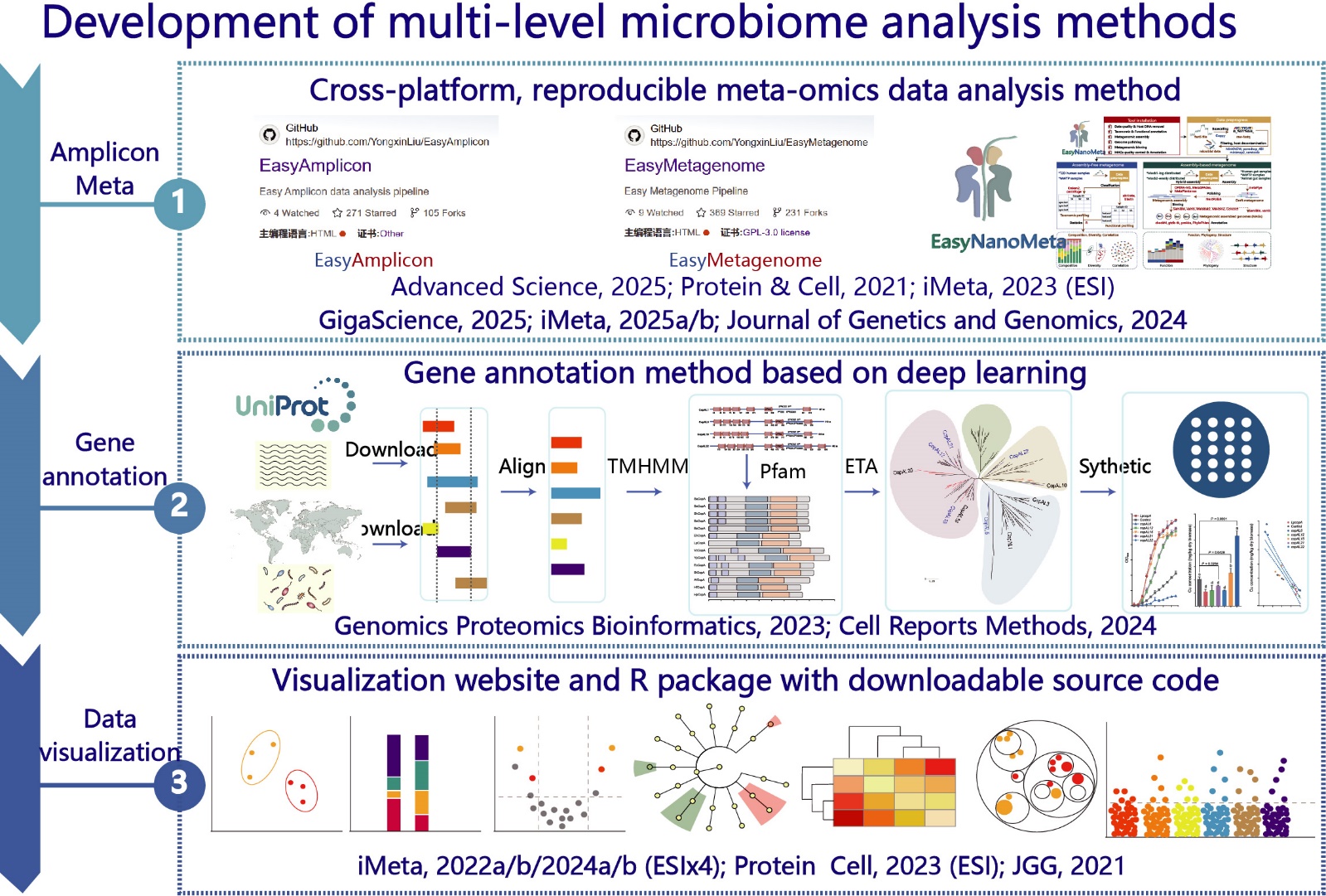
Figure 2. Development of multi-level analysis methods for microbiome.
1) High-throughput culturome enable low-cost, high-throughput identification of pure and mixed cultures; 2) Amplicon and metagenomic workflows enable precise and efficient analysis of bacterial taxonmy and functions; 3) Functional gene mining methods provide direction for experimental validation of candidates; 4) Reproducible multi-omics statistical visualization and network analysis platforms enable efficient exploration of big data.
Publications
2025
[1] Hao Luo, Defeng Bai, Zhihao Zhu, et al. 2025. EasyAmplicon 2: Expanding PacBio and Nanopore Long Amplicon Sequencing Analysis Pipeline for Microbiome. Advanced Science 12: e12447. https://doi.org/10.1002/advs.202512447
[2] Yao Wang, Junbo Yang, Huiyu Hou, et al. 2025. Advancing Plant Microbiome Research Through Host DNA Depletion Techniques. Plant Biotechnology Journal n/a: https://doi.org/10.1111/pbi.70379
[3] Zhihao Zhu, Lin Zhang, Xiaofang Yao, et al. 2025. iMeta Conference 2025: Creating high-impact international journals. iMeta 4: e70086. https://doi.org/10.1002/imt2.70086
[4] Tianyuan Zhang, Mian Jiang, Hanzhou Li, et al. 2025. Computational Tools and Resources for Long-read Metagenomic Sequencing Using Nanopore and PacBio. Genomics, Proteomics & Bioinformatics qzaf075. https://doi.org/10.1093/gpbjnl/qzaf075
[5] Defeng Bai, Tong Chen, Jiani Xun, et al. 2025. EasyMetagenome: A user-friendly and flexible pipeline for shotgun metagenomic analysis in microbiome research. iMeta 4: e70001. https://doi.org/10.1002/imt2.70001
[6] Defeng Bai, Chuang Ma, Jiani Xun, et al. 2025. MicrobiomeStatPlots: Microbiome statistics plotting gallery for meta-omics and bioinformatics. iMeta 4: e70002. https://doi.org/10.1002/imt2.70002
[7] Xiaofang Yao, Jiqiu Wu, Tengfei Ma, et al. 2025. iMeta: Boosting academic sharing and collaboration via social media. iMeta 4: e70085. https://doi.org/10.1002/imt2.70085
[8] Kai Peng, Yunyun Gao, Changan Li, et al. 2025. Benchmarking of analysis tools and pipeline development for nanopore long-read metagenomics. Science Bulletin 70: 1591-1595. https://doi.org/10.1016/j.scib.2025.03.044
[9] Kai Peng, Zeyu Tong, Changan Li, et al. 2026. Rapid reconstruction of multidrug-resistant bacterial genomes using CycloneSEQ nanopore sequencing. iMetaOmics 3: e70060. https://doi.org/10.1002/imo2.70060
[10] Yunyun Gao, Hao Luo, Hujie Lyu, et al. 2025. Benchmarking short-read metagenomics tools for removing host contamination. GigaScience 14: https://doi.org/10.1093/gigascience/giaf004
[11] Hao Luo, Yao Wang, Huiyu Hou, et al. 2025. Advances and applications in sequencing-based pathogen surveillance. aBIOTECH 100004. https://doi.org/10.1016/j.abiote.2025.100004
[12] Tao Wen, Yong-Xin Liu, Lanlan Liu, et al. 2025. ggClusterNet 2: An R package for microbial co-occurrence networks and associated indicator correlation patterns. iMeta 4: e70041. https://doi.org/10.1002/imt2.70041
[13] Yujia Wu, Lyu Hujie, Deng Xuliang, et al. 2025. Correlation between oral microbiota and dry socket at different time periods on tooth extraction. Journal of Oral Microbiology 17: 2485210. https://doi.org/10.1080/20002297.2025.2485210
[14] Yanli Li, Wenjun Zhao, Bingyan Jiang, et al. 2025. Effects of inactivated bacillus sp. DU-106 on high-fat diet-induced obesity and the gut-liver axis. International Journal of Food Science and Technology vvaf028. https://doi.org/10.1093/ijfood/vvaf028
[15] Salsabeel Yousuf, Yao Wang, Peng Yu, et al. 2025. 'Metagenomic Analysis for Unveiling Agricultural Microbiome-2nd Edition', Agronomy.
[16] Runfeng Sun, Ziheng Wang, Suyin Feng, et al. 2025. iMetaMed: Transcending Disciplinary Borders and Integrative Vision for Medicine. iMetaMed 1: e70009. https://doi.org/10.1002/imm3.70009
[17] Jingying Zhang, Bing Wang, Haoran Xu, et al. 2025. Root microbiota regulates tiller number in rice. Cell 188: 3152-3166. https://doi.org/10.1016/j.cell.2025.03.033
[18] Penghao Sun, Mengli Wang, Xuejun Chai, et al. 2025. Disruption of tryptophan metabolism by high-fat diet-triggered maternal immune activation promotes social behavioral deficits in male mice. Nature Communications 16: 2105. https://doi.org/10.1038/s41467-025-57414-4
2024
[19] Yunyun Gao, Guoxing Zhang, Shunyao Jiang, et al. 2024. Wekemo Bioincloud: A user-friendly platform for meta-omics data analyses. iMeta 3: e175. https://doi.org/10.1002/imt2.175
[20] Yunyun Gao, Kai Peng, Defeng Bai, et al. 2024. The Microbiome Protocols eBook initiative: Building a bridge to microbiome research. iMeta 3: e182. https://doi.org/10.1002/imt2.182
[21] Tianyuan Zhang, Hanzhou Li, Mian Jiang, et al. 2024. Nanopore sequencing: flourishing in its teenage years. Journal of Genetics and Genomics https://doi.org/10.1016/j.jgg.2024.09.007
[22] Chun-Lin Shi, Yong-Xin Liu. 2024. Dynamic control of lipid provisioning in plant-fungal symbiosis. Molecular Plant 367-367. https://doi.org/10.1016/j.molp.2024.02.009
[23] Yao Wang, Peng Yu, Yong-Xin Liu. 2024. Metagenomic Analysis for Unveiling Agricultural Microbiome. Agronomy 14: 981. https://doi.org/10.3390/agronomy14050981
[24] Chun-Lin Shi, Tong Chen, Canhui Lan, et al. 2024. iMetaOmics: Advancing human and environmental health through integrated meta-omics. iMetaOmics 1: e21. https://doi.org/10.1002/imo2.21
[25] Tong Chen, Yong-Xin Liu, Tao Chen, et al. 2024. ImageGP 2 for enhanced data visualization and reproducible analysis in biomedical research. iMeta 3: e239. https://doi.org/10.1002/imt2.239
[26] Mei Yang, Tong Chen, Yong-Xin Liu, et al. 2024. Visualizing set relationships: EVenn's comprehensive approach to Venn diagrams. iMeta 3: e184. https://doi.org/10.1002/imt2.184
[27] Yuanping Zhou, Yong-Xin Liu, Xuemeng Li. 2024. USEARCH 12: Open-source software for sequencing analysis in bioinformatics and microbiome. iMeta 3: e236. https://doi.org/10.1002/imt2.236
[28] Shanshan Xu, Hao Zhou, Boyang Xu, et al. 2024. Environmental microbiota and its influence on microbial succession and metabolic profiles in baijiu fermentation across three distinct-age workshops. LWT 201: 116262. https://doi.org/10.1016/j.lwt.2024.116262
[29] Shanshan Xu, Hao Zhou, Boyang Xu, et al. 2024. Deciphering layer formation in Red Heart Qu: A comprehensive study of metabolite profile and microbial community influenced by raw materials and environmental factors. Food Chemistry 451: 139377. https://doi.org/10.1016/j.foodchem.2024.139377
[30] Kai Peng, Yong-Xin Liu, Xinran Sun, et al. 2024. Large-scale bacterial genomic and metagenomic analysis reveals Pseudomonas aeruginosa as potential ancestral source of tigecycline resistance gene cluster tmexCD-toprJ. Microbiological Research 285: 127747. https://doi.org/10.1016/j.micres.2024.127747
[31] Tao Wang, Penghao Li, Xue Bai, et al. 2024. Vaginal microbiota are associated with in vitro fertilization during female infertility. iMeta 3: e185. https://doi.org/10.1002/imt2.185
[32] Ming-Hao Lv, Wen-Chong Shi, Ming-Cong Li, et al. 2024. Ms gene and Mr gene: Microbial-mediated spatiotemporal communication between plants. iMeta 3: e210. https://doi.org/10.1002/imt2.210
[33] Zhongji Pu, Chun-Lin Shi, Che Ok Jeon, et al. 2024. ChatGPT and generative AI are revolutionizing the scientific community: A Janus-faced conundrum. iMeta 3: e178. https://doi.org/10.1002/imt2.178
[34] Yao Wang, Yong-Fu Tao, Hong-Ru Wang, et al. 2024. Role of soil microbes in enhancing crop heterosis. iMetaOmics 1: e20. https://doi.org/10.1002/imo2.20
[35] Xiaofang Yao, Qiumei Liu, Yongxin Liu, et al. 2024. Managing Macadamia Decline: A Review and Proposed Biological Control Strategies. Agronomy 14: 308. https://doi.org/10.3390/agronomy14020308
[36] Shi Zhong, Yu-Qing Sun, Jin-Xi Huo, et al. 2024. The gut microbiota-aromatic hydrocarbon receptor (AhR) axis mediates the anticolitic effect of polyphenol-rich extracts from Sanghuangporus. iMeta 3: e180. https://doi.org/10.1002/imt2.180
[37] Zhiguang Qiu, Li Yuan, Chun-Ang Lian, et al. 2024. BASALT refines binning from metagenomic data and increases resolution of genome-resolved metagenomic analysis. Nature Communications 15: 2179. https://doi.org/10.1038/s41467-024-46539-7
[38] Mengli Wang, Penghao Sun, Xuejun Chai, et al. 2024. Reconstituting gut microbiota-colonocyte interactions reverses diet-induced cognitive deficits: The beneficial of eucommiae cortex polysaccharides. Theranostics 14: 4622-4642. https://doi.org/10.7150/thno.99468
[39] Min Fu, Yunhe Chen, Yong-Xin Liu, et al. 2024. Genotype-associated core bacteria enhance host resistance against kiwifruit bacterial canker. Horticulture Research uhae236. https://doi.org/10.1093/hr/uhae236

